DNA Methylation and Histone Acetylation: Definitions and Process
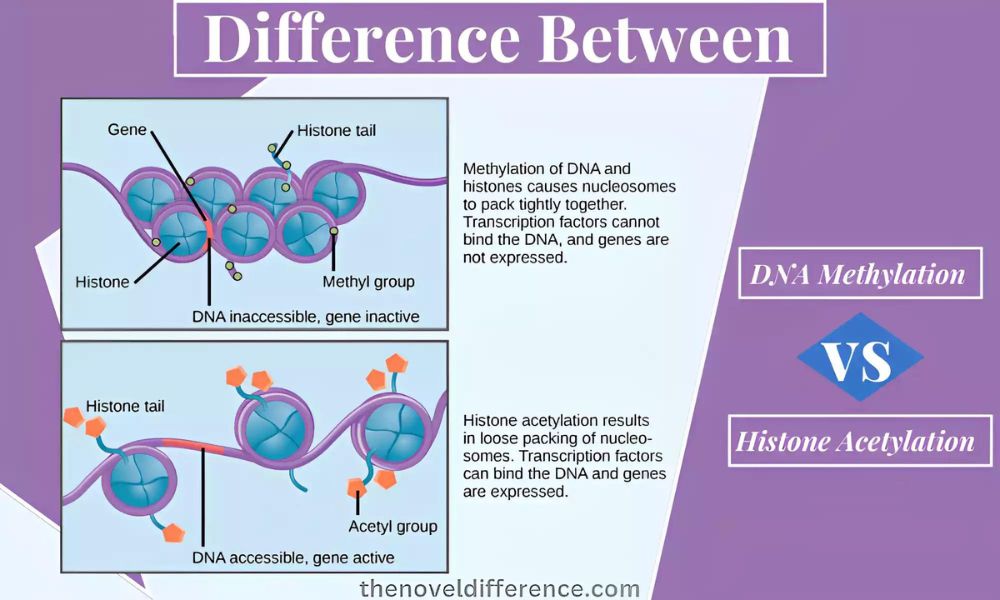
Methylation of DNA: The addition of a methyl (-CH3) group to DNA is an epigenetic change. The methylation occurs primarily at cytosine sites in the DNA sequence. Methylation occurs through DNA methyltransferases that transfer methyl groups from donor molecules onto cytosines in DNA molecules. This leads to the formation of 5-methylcytosine (also known as 5mC).
Histone Acetylation: Histone Acetylation involves adding an acetyl (-COCH3) group to the lysine residues on the tails of histone proteins. Histones are proteins that wrap DNA to form the structure known as chromatin. Histone acetylation occurs through enzymes called histone acetyltransferases that transfer acetyl groups from donor molecules – typically acetyl CoA – onto specific lysine atoms in histone tails. The lysine residue is neutralized, resulting in a relaxed chromatin structure.
DNA Methylation Process:
- Recognition: DNA Methyltransferases recognize specific DNA sequences or regions known as CpG Islands. These islands are rich in CpG Dinucleotides.
- Additions of methyl groups: DNA methyltransferases attach a methyl group on the carbon atom in the cytosine residue to produce 5-methylcytosine.
- Maintenance methylation pattern: The methylated DNA serves as a template during DNA replication. The methylation patterns of the original DNA strands are faithfully replicated onto the daughter DNA strands by DNMT1.
Histone Acetylation Process:
- Recognition: Histone Acetyltransferases recognize specific target sites in the histone tails.
- Additions of acetyl Groups: HATs add an acetyl atom from acetyl CoA to a lysine residue at the tail of the histone, leading to acetylated Lysine.
- Chromatin remodeling: By adding acetyl groups, the charge on the lysine residues is neutralized. This weakens the interaction between DNA and histones. This results in a relaxation of the chromatin structure and makes the DNA more accessible for transcription factors, regulatory proteins, etc.
Both DNA methylation and histone acetylation can be reversed. Demethylation of DNA refers to the removal of one or more methyl groups through either active enzyme-driven processes or passive degradation over time. Histone deacetylases are responsible for removing the acetyl group from histones. This results in a more compact chromatin structure.
Definition of Epigenetics
Epigenetics refers to heritable changes in gene expression or cell phenotype that occur without altering DNA sequence.This field studies various modifications and mechanisms which influence gene activity and cell function without altering DNA itself directly; such modifications might alter DNA’s physical structure itself
(i.e. histone modifications), while genetic alterations involve changing just DNA itself whereas epigenetic modifications involve alteration to its associated proteins such as histones; this can have profound impacts on gene expression by controlling their accessibility to transcription/ translation machinery located within cells themselves.
Epigenetic modifications may result from various environmental influences, including diet, stress levels, toxic exposure, or lifestyle choices.
They play key roles in the normal development and maintenance of cell identity; epigenetic modifications have even been linked with various diseases including cancer, neurological conditions, and cardiovascular ailments.
Epigenetics is a field of study that has become essential in the research world. It provides insight into how genetic and environmental information interacts to determine an organism’s phenotype and its susceptibility to disease.
Importance of epigenetic modifications in gene regulation
Epigenetic modifications play an essential role in gene regulation by altering gene accessibility to transcription and translation machinery within cells. They can have profound effects on gene expression patterns and cellular phenotypes.
Here are some key points highlighting the importance of epigenetic modifications in gene regulation:
- Development and Cellular Differentiation: Epigenetic modifications are essential for normal development and cellular differentiation. They guide the process by which a single fertilized egg gives rise to different cell types and tissues with distinct functions. Epigenetic marks regulate the activation or silencing of specific genes involved in cell fate determination, ensuring that cells differentiate into their appropriate lineages during development.
- Stability of Gene Expression Patterns: Epigenetic modifications provide stability to gene expression patterns over time and through cell divisions. They can maintain specific gene expression profiles in different cell types, enabling cells to maintain their identity and function throughout the lifetime of an organism.
- Response to Environmental Factors: Epigenetic modifications are influenced by environmental factors, such as diet, stress, toxins, and lifestyle choices. They allow organisms to adapt and respond to their environment by modulating gene expression patterns without changing the DNA sequence. This responsiveness to environmental cues can have significant implications for health, disease susceptibility, and adaptation to changing conditions.
- Gene Silencing and Activation: Epigenetic modifications, such as DNA methylation and histone modifications, can either silence or activate gene expression. DNA methylation at gene promoters often leads to gene silencing by blocking the binding of transcription factors to the DNA sequence. In contrast, certain histone modifications, like histone acetylation, promote an open chromatin structure, allowing transcription factors and other regulatory proteins to access the DNA and activate gene expression.
- Epigenetic Inheritance: Epigenetic modifications can be inherited from one generation to the next, independent of changes in the DNA sequence. This phenomenon, known as epigenetic inheritance, can influence the expression of genes and phenotypic traits in offspring. It provides a mechanism for transmitting acquired epigenetic changes, including those induced by environmental factors, across generations.
- Disease Development and Progression: Aberrant epigenetic modifications have been associated with various diseases, including cancer, neurological disorders, cardiovascular diseases, and metabolic disorders. Altered epigenetic regulation can lead to abnormal gene expression patterns, disrupt cellular homeostasis, and contribute to disease initiation and progression. Understanding the role of epigenetic modifications in disease processes offers opportunities for developing novel therapeutic strategies.
Epigenetic modifications are critical for regulating gene expression and maintaining cellular identity. They allow cells to respond to environmental cues, ensure proper development and differentiation, and contribute to the stability and adaptability of gene expression patterns. Dysfunction in epigenetic regulation can have profound implications for health and disease.
DNA Methylation
DNA methylation is an epigenetic alteration that occurs when methyl groups (-CH3) are added to DNA molecules. It has been studied for many years as one of the epigenetic modifications’ hallmarks.
DNA methylation typically occurs at CpG sites where cytosines are immediately followed by guanine nucleotides in sequence; specifically at Cytosine-Phenylalanine G (CPG) sites.
DNA methylation occurs through enzymes known as DNA methyltransferases (DNMTs), which transfer methyl groups from donor molecules such as S-adenosyl methionine (SAM) directly onto carbon atoms found on cytosine residues to form 5-methylcytosine (5mC).
Early development establishes DNA methylation patterns that remain constant over the lifetime of an organism. Maintenance of these methylation patterns is performed primarily through maintenance of DNA methyltransferase DNMT1, ensuring that any original parent strand methylation patterns are precisely copied onto newly synthesized daughter strands during DNA replication.
DNA methylation plays an essential part in gene regulation by altering how accessible genes are to transcriptional machinery. DNA methylation at promoter regions often results in reduced gene expression or silencing and may physically prevent transcription factors and regulatory proteins from binding with DNA sequences, thus stopping transcription from the beginning.
As well as silencing genes, DNA methylation also plays an essential part in gene regulation by altering chromatin structure and organization. Methylated DNA attracts proteins that modify histones for greater compactness and transcriptionally repressive chromatin structure.
While DNA methylation has often been linked with gene silencing, its impact can be reversed at any point through active demethylation or through passive effects over time as cell division reduces maintenance of DNMT activity and its efficiency declines.
Environmental factors, including diet, stress levels, and environmental toxins, may impact DNA methylation patterns; changes can have major ramifications on gene expression patterns, cell functions and development, and disease susceptibility.
Understanding DNA methylation’s impact on gene regulation is paramount to comprehending complex mechanisms underlying normal growth, differentiation, and disease susceptibility.
Histone Acetylation
Histone acetylation is an epigenetic modification that adds an acetyl group (-COCH3) to histone tail lysines for epigenetic modification of DNA wrapped up by histone proteins forming the structure known as chromatin.
Histone acetylation occurs via enzymes called histone acetyltransferase. HATs transfer an acetyl group from donor molecules such as acetyl-CoA onto specific lysine residues on histone tails to neutralize positive charges that arise as part of this process, creating more relaxed chromatin structures and providing for cell division and proliferation.
Histone acetylation plays an essential role in gene regulation. Usually associated with transcriptional activation or increased gene expression, histone acetylation changes chromatin structure by loosening DNA-histone interactions to make DNA more accessible for transcription.
Acetylation of histones creates binding sites for bromodomain-containing proteins to bind. Once bound, they recruit additional factors like transcriptional activators, coactivators, and chromatin remodelers in order to facilitate gene transcription; together they promote the assembly of transcriptional complexes that initiate and enhance gene expression.
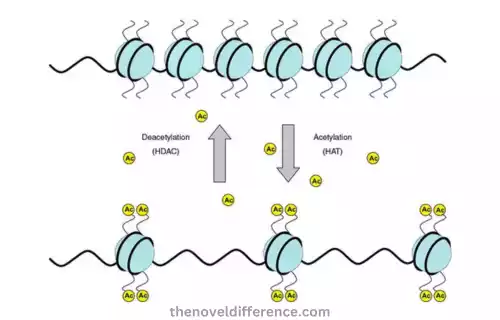
Histone deacetylases (HDACs), however, play an opposite role and remove acetyl groups from histones through deacetylation processes that stimulate compression of chromatin structure and transcriptional repression.
Histone acetylation is an ever-evolving process influenced by environmental conditions, signaling pathways, and cell processes. Achieve balance between histone acetylation and deacetylation plays a pivotal role in controlling gene expression patterns as well as maintaining cell health homeostasis.
Histone acetylation and histone modifications such as phosphorylation and ubiquitination create an intricate regulatory network to control gene transcriptional states and cell functions related to disease or development processes.
Investigating histone acetylation and its effect on chromatin structure and gene regulation provides invaluable insights into epigenetic regulatory mechanisms. This has implications for normal development, cancer, neurological disorders, and cardiovascular diseases among others.
Differences between DNA Methylation and Histone Acetylation
DNA Methylation & Histone Acetylation play different epigenetic roles that are important in gene regulation.
Here are some key differences between histone acetylation and DNA methylation:
- The mechanism for Modification:
-
- DNA Methylation: Addition of a methyl (-CH3) group to DNA molecules, specifically in CpG sites.
- Histone Acetylation: Addition of an acetyl (-COCH3) group to lysine residues in histone proteins that are associated with DNA.
- Target Moleculars:
-
- DNA Methylation: This occurs directly on DNA molecules themselves, specifically at cytosine methyl residues.
- Histone Acetylation: Occurs in the histone proteins around which DNA wraps, altering the accessibility and structure of chromatin.
- Stability:
-
- DNA Methylation: This is generally stable and heritable across cell divisions as DNA methyltransferases maintain it during replication.
- Histone Acetylation: More Dynamic and Reversible. Acetyl groups are added or removed quickly, allowing rapid changes in gene transcription.
- Effects of gene expression:
-
- DNA Methylation: is Often linked to gene silencing and reduced gene expression. Methylation of promoter regions may physically prevent transcription by inhibiting transcription factors from binding.
- Histone Acetylation: Is generally associated with increased gene expression or gene activation. Acetylation promotes a relaxed chromatin structure that allows transcription factors and other regulatory proteins better access to DNA.
- Interplay & cooperation:
-
- Histone Acetylation and DNA Methylation: Histone acetylation and DNA methylation can affect each other’s effects and patterns. DNA methylation, for example, can recruit proteins to modify histones. This leads to a compacter and transcriptionally repressive structure of chromatin. Histone acetylation also influences the binding of proteins that are involved in DNA demethylation and methylation.
Understanding how DNA methylation versus histone acetylation contributes to gene regulation, cell processes, and disease development can provide invaluable insight. These epigenetic changes interact to maintain cell identity and shape gene expression patterns.
What are the similarities between DNA Methylation and Histone Acetylation?
Although DNA methylation is a distinct epigenetic modification, it shares some similarities with histone acetylation in terms of their roles and consequences.
Here are some similarities between histone acetylation and DNA methylation:
- Regulation of gene expression: DNA methylation as well as histone acetylation play a role in the regulation. They can influence the accessibility of genes for the transcriptional machinery and thereby affect whether a particular gene is turned off or on.
- Epigenetic Modifications: DNA-methylation and histone-acetylation can cause stable, heritable and permanent changes in gene expression.
- Dynamic, reversible, and dynamic: While DNA methylation tends to be more stable than histone-acetylation in general, both modifications are reversible and dynamic. These modifications can be removed or added by enzymatic means, which allows for flexible gene regulation to respond to environmental cues and developmental changes.
- Interplay and cooperation: DNA-methylation patterns and effects can be affected by each other. DNA methylation, for example, can recruit proteins to modify histones and lead to changes in the chromatin structure. Histone acetylation, on the other hand, can affect the binding of proteins that are involved in DNA demethylation and methylation.
- Impact of both modifications on chromatin: Both changes can affect the structure of the chromatin complex, which is made up of DNA and histone protein. DNA methylation or histone acetylation may affect the compactness of chromatin and the accessibility of the chromatin. This can influence the binding of transcription factors and regulatory proteins to the DNA.
- Role during development and disease: Both DNA methylation and histone acetylation, play an important role in the normal development of a child and in various diseases. Dysregulation can cause aberrant gene expression and lead to diseases such as cancer, neurological disorders, and cardiovascular disease.
Understanding the similarities between histone acetylation and DNA methylation helps us understand the interconnectedness of epigenetic mechanisms and their impact on cell processes and disease states.
DNA Methylation vs Histone Acetylation in Tabular Form
Sure! Here’s a tabular form highlighting the key differences between DNA methylation and histone acetylation:
DNA MethylationHistone Acetylation
| Mechanism | Addition of a methyl group (-CH3) to cytosine residues in CpG sites of DNA | Addition of an acetyl group (-COCH3) to lysine residues on histone proteins |
| Target molecules | DNA | Histone proteins associated with DNA |
| Stability | Generally stable and heritable over cell divisions | More dynamic and reversible |
| Effects on gene expression | Gene silencing or reduced gene expression | Gene activation or increased gene expression |
| Impact on chromatin structure | This can lead to a more compact and transcriptionally repressive chromatin structure | Results in a more relaxed and transcriptionally active chromatin structure |
| Interplay and cooperation | Can recruit proteins that modify histones, affecting chromatin structure | Can influence the binding of proteins involved in DNA methylation and demethylation |
| Role in gene regulation | Inhibits transcription initiation by blocking the binding of transcription factors | Facilitates transcription initiation by promoting accessibility to transcriptional machinery |
| Environmental influence | Can be influenced by various factors such as diet, stress, toxins, and lifestyle choices | Can be influenced by external signals and cellular processes |
| Role in development and diseases | Plays a critical role in normal development and diseases such as cancer, neurological disorders, and cardiovascular diseases | Also crucial in the development and implicated in various diseases |
| Examples of enzymes involved | DNA methyltransferases (DNMTs) | Histone acetyltransferases (HATs) and histone deacetylases (HDACs) |
Even though DNA methylation, histone acetylation, and their interactions and cooperation play key roles in gene regulation as well as contributing to disease states or cell processes, there can still be differences.
Conclusion
DNA methylation and histone acetylation are two interdependent epigenetic modifications that serve an integral part in gene regulation. DNA methylation involves attaching methyl groups to specific cytosine residues within DNA at CpG sites to silence genes; while histone acetylation adds an acetyl group directly onto histone proteins resulting in more open chromatin structures for increased gene expression.
Both DNA methylation and histone acetylation modify histone proteins that wrap DNA molecules. Both modifications can influence whether genes can access transcriptional machinery for expression purposes; and can even act in tandem to shape each other’s effects, with DNA methylation impacting histone acetylation having an influence over it, and vice versa.
DNA methylation tends to be stable across cell divisions while histone acetylation tends to be dynamic and reversible; both modifications can be affected by environmental influences that influence normal development, differentiation processes, and disease processes; their dysregulation has also been implicated in various conditions like cancer, neurological diseases, and cardiovascular illnesses.