Introduction of hnrna and mrna
Gene expression is a complex phenomenon that involves various stages and molecules. Among these molecules, hnrna (heterogeneous nuclear rna) and mrna (messenger rna) play vital roles in the synthesis of proteins. While both types of rna are involved in gene expression, there are significant differences between hnrna and mrna. In this article, we will delve into these dissimilarities, exploring their structures, functions, and implications.
Importance of hnrna and mrna in gene expression
Hnrna (heterogeneous nuclear rna) and mrna (messenger rna) play crucial roles in gene expression, which is the process by which genetic information is used to produce functional proteins.
The importance of hnrna and mrna in gene expression can be understood through the following points:
1. Transcription: hnrna serves as the precursor to mrna during the process of transcription. Transcription is the first step in gene expression, where the dna sequence is copied into hnrna by rna polymerase. This initial transcription allows for the synthesis of hnrna molecules, which are larger and more heterogeneous than mature mrna.
2. Rna processing: hnrna undergoes post-transcriptional modifications, including capping, polyadenylation, and splicing, to produce mature mrna. These modifications are essential for mrna stability, transport, and translation efficiency. The addition of a 5′ cap and a poly-a tail to mrna protects it from degradation and assists in its recognition and transport out of the nucleus.
3. Alternative splicing: hnrna contains both exons (coding regions) and introns (non-coding regions). Splicing, a process unique to hnrna, removes the introns and joins the exons together to form mature mrna. Alternative splicing allows for the generation of multiple mrna isoforms from a single hnrna precursor, thereby increasing the diversity of proteins that can be produced from a single-gene.
4. Mrna stability: mrna is typically more stable than hnrna, allowing it to persist in the cytoplasm for translation. The modifications of the 5′ cap and poly-a tail play crucial roles in mrna stability. The length of the poly-a tail influences mrna degradation rates, thereby regulating gene expression levels.
5. Translation: mrna serves as the template for protein synthesis during translation. Ribosomes recognize the start codon on mrna and proceed to read the codons, translating them into specific amino acids. The information encoded in mrna determines the amino acid sequence of the resulting protein.
6. Gene regulation: the levels of hnrna and mrna can be regulated at both transcriptional and post-transcriptional levels. Regulatory proteins, transcription factors, and epigenetic modifications can control the transcription of hnrna, thereby modulating gene expression. Post-transcriptional regulation includes mechanisms such as rna interference (rnai) and micrornas, which can degrade or inhibit the translation of specific mrna molecules, influencing protein expression.
7. Disease implications: dysregulation of hnrna and mrna can lead to various disease conditions. Aberrant splicing, mutations, or altered expression levels of mrna can result in abnormal protein production, contributing to genetic disorders, developmental abnormalities, or diseases like cancer. Understanding the mechanisms and importance of hnrna and mrna in gene expression is critical for studying and potentially treating these diseases.
Hnrna and mrna are key players in gene expression, regulating the synthesis of proteins in cells. Their roles encompass transcription, processing, stability, translation, and gene regulation. Investigating hnrna and mrna provides valuable insights into the mechanisms underlying gene expression and its impact on cellular functions and disease processes.
Structure and processing of hnrna
The structure and processing of hnrna (heterogeneous nuclear rna) are crucial steps in gene expression. Hnrna serves as the precursor molecule to mrna (messenger rna) and undergoes several modifications before it can be transformed into mature mrna.
Here is an overview of the structure and processing of hnrna:
1. Structure of hnrna:
• hnrna is a heterogeneous mixture of rna molecules present in the nucleus of eukaryotic cells.
• it is synthesized through the process of transcription, where rna polymerase binds to the dna template and synthesizes a complementary rna strand.
• hnrna molecules are typically larger and more heterogeneous than mature mrna.
• hnrna contains both exons (coding regions) and introns (non-coding regions).
2. Transcription of hnrna:
• transcription begins with the binding of rna polymerase to the promoter region of a gene on the dna template strand.
• rna polymerase synthesizes an rna strand complementary to the dna template strand, incorporating nucleotides according to the base-pairing rules (a with u, g with c, etc.).
• transcription results in the synthesis of a primary hnrna transcript that contains both exonic and intronic sequences.
3. Post-transcriptional modifications of hnrna:
• after transcription: hnrna undergoes several processing steps to become mature mrna. These modifications are essential for mrna stability, transport, and efficient translation.
A. Addition of poly-a tail:
• a poly-a tail: consisting of multiple adenine nucleotides, is added to the 3′ end of the hnrna molecule.
• the poly-a tail aids in mrna stability: protects it from degradation and assists in its transport from the nucleus to the cytoplasm.
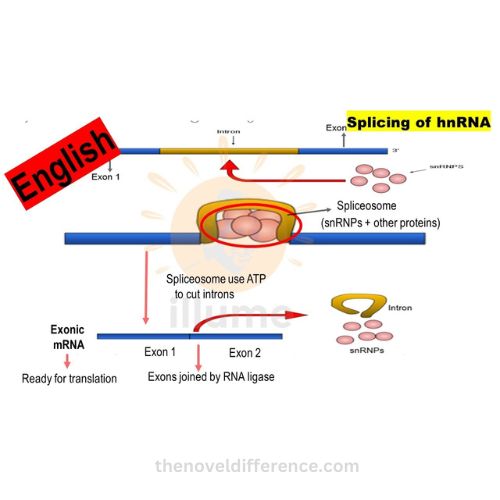
B. Splicing of introns:
• splicing is a crucial step in hnrna processing that removes introns (non-coding regions) and joins exons (coding regions) together.
• splicing is carried out by the spliceosome, a complex composed of small nuclear ribonucleoproteins (snrnps) and other proteins.
• the spliceosome recognizes specific sequences at the boundaries of introns, cuts them out, and joins the adjacent exons to form a mature mrna molecule.
• alternative splicing can generate multiple mrna isoforms from a single hnrna precursor, increasing protein diversity.
4. Mature mrna formation:
• after the post-transcriptional modifications, hnrna is processed into mature mrna.
• the mature mrna contains only the exonic sequences that code for proteins, with the introns removed.
• the mature mrna is then transported from the nucleus to the cytoplasm, where it can be translated into a functional protein.
The processing of hnrna into mature mrna is a highly regulated and intricate process. It ensures that only the necessary coding regions are retained, and the mrna molecule is modified to ensure stability, transport, and efficient translation. These modifications are crucial for the proper functioning of gene expression and protein synthesis.
Addition of 5′ cap
The addition of a 5′ cap is a post-transcriptional modification that occurs during the processing of hnrna (heterogeneous nuclear rna) to mature mrna (messenger rna). The 5′ cap is a modified guanosine nucleotide (m7g) that is added to the 5′ end of the nascent rna molecule.
Here’s an overview of the addition of the 5′ cap:
1. Capping enzymes recognition:
• shortly after transcription begins, as the rna polymerase synthesizes the hnrna transcript, capping enzymes recognize specific sequences and structures near the 5′ end of the nascent rna.
• the recognition of these signals triggers the capping process.
2. Addition of guanosine triphosphate (gtp):
• the first step of the capping process involves the addition of a guanosine triphosphate molecule (gtp) in a reverse orientation to the 5′ end of the nascent rna.
• the gtp is linked to the rna molecule through a 5′-5′ triphosphate bridge, rather than the typical 5′-3′ phosphodiester bond.
3. Methylation of the guanosine residue:
• after the gtp is added, the guanosine residue undergoes a methylation process.
• a methyl group is added to the 7th position of the guanosine base, resulting in the modified guanosine nucleotide, m7g.
4. Linkage between the cap and the rna:
• once the m7g cap is formed, it is covalently linked to the 5′ end of the rna molecule.
• the cap is connected to the nascent rna through a unique 5′-5′ triphosphate linkage.
• this linkage protects the nascent rna from degradation and helps stabilize the rna molecule.
The addition of the 5′ cap serves several important functions:
1. Mrna stability: the cap structure protects the mrna from degradation by exonucleases, enzymes that degrade rna from the ends. The cap acts as a physical barrier, preventing the exonucleases from reaching the rna molecule.
2. Mrna recognition and transport: the cap plays a crucial role in the recognition and binding of the mrna by the cellular machinery involved in mrna processing, export from the nucleus, and translation in the cytoplasm. It acts as a recognition signal for various proteins involved in these processes.
3. Translation initiation: the cap structure is recognized by the translation initiation complex during protein synthesis. It assists in the proper positioning of the ribosome on the mrna, facilitating the efficient initiation of translation.
The addition of the 5′ cap is an essential modification that occurs early in the processing of hnrna and is critical for the stability, recognition, and efficient translation of mature mrna.
Structure and processing of mrna
The structure and processing of mrna (messenger rna) involve various post-transcriptional modifications that are crucial for its stability, transport, and efficient translation into protein.
Here is an overview of the structure and processing of mrna:
1. Structure of mrna:
• mrna is a single-stranded rna molecule that carries genetic information from the dna to the ribosomes for protein synthesis.
• it consists of three main regions:
A. 5′ untranslated region (5′ utr): located at the 5′ end of the mrna molecule, this region contains regulatory elements and binding sites for various proteins involved in translation initiation.
B. Coding region: this region, also known as the open reading frame (orf), contains the nucleotide sequence that encodes the amino acid sequence of the protein.
C. 3′ untranslated region (3′ utr): found at the 3′ end of the mrna, this region contains regulatory elements involved in mrna stability, localization, and translational control.
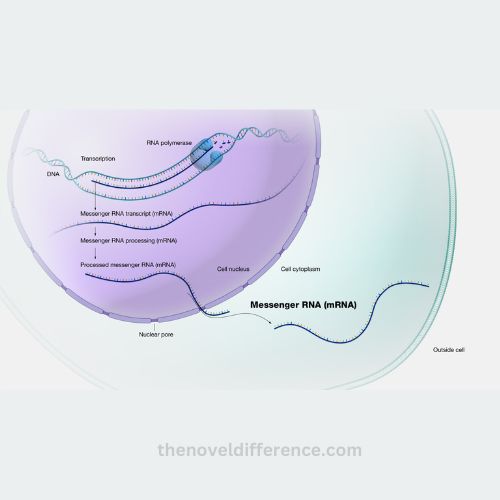
2. Transcription of mrna:
• transcription is the process by which rna polymerase synthesizes an rna molecule complementary to the dna template.
• during transcription, rna polymerase binds to the promoter region of a gene and initiates the synthesis of pre-mrna (hnrna).
• the pre-mrna molecule undergoes subsequent processing steps to become mature mrna.
3. Post-transcriptional modifications of mrna:
• after transcription, pre-mrna undergoes several modifications to become mature mrna. These modifications occur in the nucleus before the mrna is transported to the cytoplasm for translation.
A. Addition of poly-a tail:
• a poly-a tail, consisting of multiple adenine nucleotides, is added to the 3′ end of the pre-mrna.
• the poly-a tail helps stabilize the mrna, protects it from degradation, and plays a role in mrna export from the nucleus.
B. Alternative splicing:
• alternative splicing is a process in which different combinations of exons and introns are selected, resulting in the production of multiple mrna isoforms from a single pre-mrna.
• it allows for the generation of different protein variants from a single gene and increases proteomic diversity.
4. Mrna export:
• mature mrna molecules are transported from the nucleus to the cytoplasm through nuclear pores.
• export factors and mrna-binding proteins facilitate the export process, ensuring that only fully processed and functional mrna molecules are transported.
5. Translation:
• mature mrna serves as a template for protein synthesis during translation.
• ribosomes recognize the 5′ cap and scan the mrna molecule until they find the start codon (typically aug) within the coding region.
• the ribosome then reads the codons in the mrna, translating them into specific amino acids to synthesize the protein.
The structure and processing of mrna are essential for proper gene expression and protein synthesis. The modifications, such as the addition of the 5′ cap and poly-a tail, alternative splicing, and mrna export, ensure mrna stability, localization, and translational efficiency. These processes play crucial roles in regulating gene expression and the production of functional proteins.
Addition of 5′ cap
The addition of a 5′ cap is a post-transcriptional modification that occurs during the processing of mrna (messenger rna) in eukaryotic cells. It involves the addition of a modified guanosine nucleotide (m7g) to the 5′ end of the mrna molecule.
Here is an overview of the process of 5′ capping:
1. Capping enzyme recruitment:
• shortly after transcription initiation, when rna polymerase has synthesized a short rna sequence, a capping enzyme complex is recruited to the growing mrna molecule.
• the capping enzyme complex recognizes specific signals on the rna and interacts with rna polymerase to initiate the capping process.
2. Guanosine triphosphate (gtp) addition:
• the capping enzyme catalyzes the addition of a gtp molecule to the 5′ end of the nascent mrna molecule.
• the gtp is added in a reverse orientation, with the 5′ end of the gtp linked to the 5′ end of the mrna molecule through a unique 5′ to 5′ triphosphate linkage.
3. Guanine methylation:
• following the addition of the gtp, another enzymatic reaction occurs where a methyl group is added to the guanine base of the attached gtp.
• this methyl group is derived from s-adenosyl methionine (sam), a common methyl donor in cellular processes.
4. Additional methylations (optional):
• in some cases, additional methyl groups may be added to the first transcribed nucleotides of the mrna molecule, creating a “cap 1” structure.
• these additional methylations are usually present in more complex organisms and may provide further protection and regulation of mrna.
The addition of a 5′ cap to mrna serves several important functions:
1. Mrna stability: the 5′ cap protects the mrna from degradation by exonucleases, which can degrade rna from the 5′ end. The cap structure provides stability to the mrna molecule, prolonging its lifespan in the cytoplasm.
2. Facilitating translation initiation: the 5′ cap plays a vital role in the recognition and binding of the ribosome during translation initiation. It helps in the assembly of the translation machinery, promoting efficient translation of the mrna.
3. Mrna export: the presence of the 5′ cap is recognized by proteins involved in mrna export from the nucleus to the cytoplasm. The cap structure facilitates the transport of mrna through nuclear pores.
4. Regulation of gene expression: the 5′ cap is involved in the regulation of gene expression. It can influence the rate of mrna degradation, the efficiency of translation initiation, and the interaction of mrna with regulatory proteins.
The addition of a 5′ cap to mrna during post-transcriptional processing is a critical step in mrna maturation. The 5′ cap protects the mrna from degradation, aids in translation initiation, facilitates mrna export, and contributes to the regulation of gene expression.
Differences between hnrna and mrna
Heterogeneous nuclear rna (hnrna) and messenger rna (mrna) are two distinct forms of rna involved in gene expression, with some notable differences between them.
Here are the key differences between hnrna and mrna:
1. Structure and composition:
• hnrna: heterogeneous nuclear rna is the primary transcript of a gene synthesized during transcription. It contains both coding regions (exons) and non-coding regions (introns). Hnrna is larger in size and more heterogeneous due to the presence of introns and other non-coding sequences.
• mrna: messenger rna is the mature rna molecule that is produced after the processing of hnrna. It typically consists of only exonic sequences that encode the information for protein synthesis. Mrna is smaller in size and more homogeneous compared to hnrna.
2. Processing:
• hnrna: heterogeneous nuclear rna undergoes various post-transcriptional modifications, including capping, polyadenylation, and splicing. These modifications are necessary to convert hnrna into mature mrna.
• mrna: messenger rna undergoes post-transcriptional processing, which includes the removal of introns through splicing and the addition of a 5′ cap and a poly-a tail. These modifications stabilize the mrna, facilitate its transport from the nucleus to the cytoplasm, and regulate its translation.
3. Stability and lifespan:
• hnrna: heterogeneous nuclear rna is relatively unstable and has a short lifespan. It is rapidly degraded in the nucleus if not properly processed into mature mrna.
• mrna: messenger rna is more stable and has a longer lifespan compared to hnrna. The addition of a 5′ cap and a poly-a tail protects the mrna from degradation by exonucleases, ensuring its longevity for translation.
4. Function:
• hnrna: heterogeneous nuclear rna serves as an intermediate molecule during gene expression. It undergoes processing to generate mature mrna, which carries genetic information from dna to ribosomes for protein synthesis.
• mrna: messenger rna is responsible for carrying genetic information from the nucleus to the ribosomes in the cytoplasm. It serves as a template for protein synthesis during translation.
5. Localization:
• hnrna: heterogeneous nuclear rna is predominantly located in the nucleus of eukaryotic cells, where transcription occurs.
• mrna: messenger rna is translocated from the nucleus to the cytoplasm, where it associates with ribosomes for translation.
Hnrna and mrna represent different stages of rna processing and have distinct structural and functional characteristics. Hnrna is the initial primary transcript synthesized during transcription, while mrna is the processed and mature rna molecule that carries the genetic information for protein synthesis. The processing steps, stability, function, and localization differ between hnrna and mrna, highlighting their respective roles in gene expression.
Regulation of hnrna and mrna
Regulation of hnrna (heterogeneous nuclear rna) and mrna (messenger rna) plays a crucial role in controlling gene expression and ensuring proper cellular functions. The regulation of both types of rna occurs at multiple levels, including transcriptional, post-transcriptional, and translational regulation.
Here is an overview of the regulation of hnrna and mrna:
1. Transcriptional regulation:
• transcriptional regulation controls the synthesis of hnrna, which is the precursor for mrna.
• transcription factors bind to specific dna sequences in the promoter regions of genes, either enhancing (activators) or inhibiting (repressors) transcription.
• regulatory proteins can influence the recruitment of rna polymerase to the promoter, thereby modulating the initiation of transcription.
• epigenetic modifications, such as dna methylation and histone modifications, can also impact gene transcription by altering the accessibility of the dna to transcriptional machinery.
2. Post-transcriptional regulation:
• post-transcriptional: Regulation occurs after hnrna synthesis and involves various processes that control mrna maturation, stability, and localization.
• splicing: alternative splicing can generate different mrna isoforms by selectively removing introns and combining exons. This process allows for increased protein diversity from a single-gene.
• rna processing: the addition of a 5′ cap and a poly-a tail to the mrna molecule influences stability, mrna export, and translation efficiency.
• mrna stability: regulatory proteins and non-coding rnas, such as micrornas (mirnas) and long non-coding rnas (lncrnas), can bind to mrna molecules and modulate their stability. This regulation can affect mrna degradation rates and ultimately protein expression levels.
3. Translational regulation:
• translational regulation controls the efficiency of protein synthesis from mrna molecules.
• regulatory proteins and small regulatory rnas can bind to specific regions of mrna, either inhibiting or promoting translation initiation.
• the presence or absence of specific sequences in the mrna molecule can affect the accessibility of the ribosome to the mrna, influencing translation efficiency.
• post-translational modifications of regulatory proteins can impact their activity, thereby modulating translation.
4. Feedback regulation:
• the expression of hnrna and mrna can be regulated by feedback mechanisms involving the proteins they encode.
• some proteins produced from mrna molecules can act as transcription factors or regulatory proteins that modulate the expression of their own genes or genes involved in their synthesis.
• feedback loops can help maintain the balance of gene expression and regulate cellular processes.
The regulation of hnrna and mrna is a complex and dynamic process that involves multiple levels of control. Transcriptional, post-transcriptional, translational, and feedback mechanisms work together to fine-tune gene expression, allowing cells to respond to internal and external cues and maintain proper cellular functions.
Conclusion
Understanding the difference between hnrna and mrna is crucial for comprehending the intricate process of gene expression. While hnrna serves as the precursor molecule, containing both exons and introns, mrna is the mature and processed molecule that carries the genetic information for protein synthesis. The removal of introns and other modifications during rna processing transforms hnrna into mrna, making it ready for translation.
These distinctions between hnrna and mrna highlight their respective roles in gene expression and emphasize their significance in various cellular processes. Expanding our knowledge of these differences deepens our understanding of the complexities of genetic regulation.
