CGH and Array CGH use microarrays to detect and analyze genomic alterations. It provides a higher resolution and a more comprehensive assessment of the genome compared to traditional CGH methods.
Array CGH enables the simultaneous evaluation of thousands to millions of DNA loci or probes across the genome, allowing for a detailed analysis of copy number variations (CNVs), gene dosage alterations, and other structural changes.
Array CGH involves the use of a DNA microarray chip containing immobilized genomic DNA fragments or probes that cover specific regions of the genome.
These probes are designed to target known genes, regulatory regions, and other functionally relevant genomic sequences. The patient’s DNA and a reference DNA sample (often a healthy individual’s DNA) are labeled with different fluorophores, typically Cy5 and Cy3, respectively.
Importance of studying genomic alterations
Studying genomic alterations is of paramount importance in various fields of research and clinical practice.
Here are some key reasons highlighting the significance of investigating genomic alterations:
- Disease Understanding and Diagnosis: Genomic alterations play a fundamental role in the development and progression of various diseases, including cancer, genetic disorders, and complex diseases. Researchers can gain more insight into the molecular processes behind diseases by investigating genomic changes. They can also identify genes that cause disease and better understand the pathways involved. This knowledge contributes to accurate disease diagnosis, prognosis, and treatment selection.
- Personalized Medicine: Genomic alterations provide crucial information for personalized medicine approaches. Clinicians can customize treatment plans by analyzing a patient’s genome profile. This includes copy number variations, mutations, and structural rearrangements. This approach improves treatment efficacy, minimizes adverse effects, and optimizes patient outcomes.
- Targeted Therapy: Genomic alterations serve as potential therapeutic targets. Identifying specific genetic abnormalities allows researchers and clinicians to develop targeted therapies that directly address the underlying molecular alterations driving the disease. For example, in cancer treatment, targeted therapies designed to inhibit specific oncogenes or pathways associated with genomic alterations have shown remarkable success in improving patient outcomes.
- Early Detection and Prevention: Genomic alterations can serve as biomarkers for early detection and prevention strategies. By identifying genetic changes associated with disease predisposition or early stages of disease development, individuals at risk can undergo appropriate screening, surveillance, and preventive interventions. Early detection increases the chances of successful intervention and improves overall patient outcomes.
- Understanding Disease Heterogeneity: Genomic alterations contribute to the heterogeneity observed within diseases. Different patients with the same clinical diagnosis can demonstrate different genomic alterations, leading to differences in disease severity, treatment response, and prognosis. Studying genomic alterations helps elucidate the underlying molecular heterogeneity, leading to improved patient stratification, targeted therapies, and personalized treatment approaches.
- Evolutionary Studies and Comparative Genomics: Genomic alterations provide insights into evolutionary processes and comparative genomics. By comparing genomes across species, researchers can identify conserved regions and evolutionary breakpoints, and understand the genetic basis of phenotypic variations. This knowledge aids in deciphering evolutionary relationships, understanding the functional significance of genomic elements, and identifying potential targets for further research.
- Genetic Counseling and Family Planning: Studying genomic alterations is crucial for genetic counseling and family planning. Individuals and their families can make more informed decisions regarding family planning, reproductive options, and genetic risk assessment by recognizing disease-causing mutations or chromosomal abnormalities that inform them about familial risks. This empowers individuals to manage their genetic health and make choices that align with their personal values and goals.
Studying genomic alterations has far-reaching implications for disease understanding, diagnosis, personalized medicine, targeted therapy, early detection, and prevention.
It paves the way for advancements in precision medicine, genetic counseling, and the development of innovative therapeutic approaches. Researchers and clinicians are able to improve patient outcomes by unraveling the complexity of genomic alterations. It will eventually lead to a greater understanding of human health.
Comparative Genomic Hybridization (CGH)
Comparative Genomic Hybridization (CGH) is a molecular cytogenetic technique used to detect and analyze genomic alterations on a global scale. Provides a genome-wide snapshot of copy number variations (CNVs), copy amplifications, deletions, and structural DNA modifications across the entire genome.
The principle of CGH involves comparing the relative DNA copy number changes between a test sample (patient DNA) and a control sample (reference DNA).
The DNA from both samples is labeled with different fluorophores, typically Cy5 and Cy3, respectively. The labeled DNA samples are then co-hybridized to normal metaphase chromosomes or DNA microarrays.
The labeled patient and reference DNA are hybridized into metaphase chromosomes. The DNA from the test and control samples compete for binding to the patient’s metaphase chromosomes.
The fluorescence signals emitted by the labeled DNA are visualized and analyzed to identify differences in signal intensity along the chromosomes. An increase or decrease in fluorescence intensity indicates a gain or loss of genetic material, respectively.
With the development of microarray technology, Array CGH emerged as an advanced version of CGH. Array CGH utilizes a DNA microarray chip containing immobilized genomic DNA fragments or probes.
The microarray chip is designed to cover specific regions of the genome, including known genes, regulatory regions, and other functionally relevant sequences. The labeled patient and reference DNA samples are co-hybridized to the microarray, where they competitively bind to the immobilized probes.
Following hybridization, the microarray is scanned to detect the fluorescence intensities of the bound labeled DNA. The fluorescence signals are quantified and analyzed to determine the relative copy number changes between the patient and reference DNA.
By comparing the signal ratios, researchers can identify gains or losses of genetic material, indicating genomic alterations such as amplifications or deletions.
The resolution of Array CGH is determined by the density and coverage of probes on the microarray, allowing for a more detailed analysis of genomic alterations compared to traditional CGH.
CGH techniques, including traditional CGH and Array CGH, have been widely used in various research and clinical applications. They have played an essential part in identifying genomic changes associated with cancer, genetic disorders, and other forms of illness.
CGH provides valuable information for diagnostic purposes, prognostic assessment, treatment decision-making, and understanding the genetic basis of diseases.
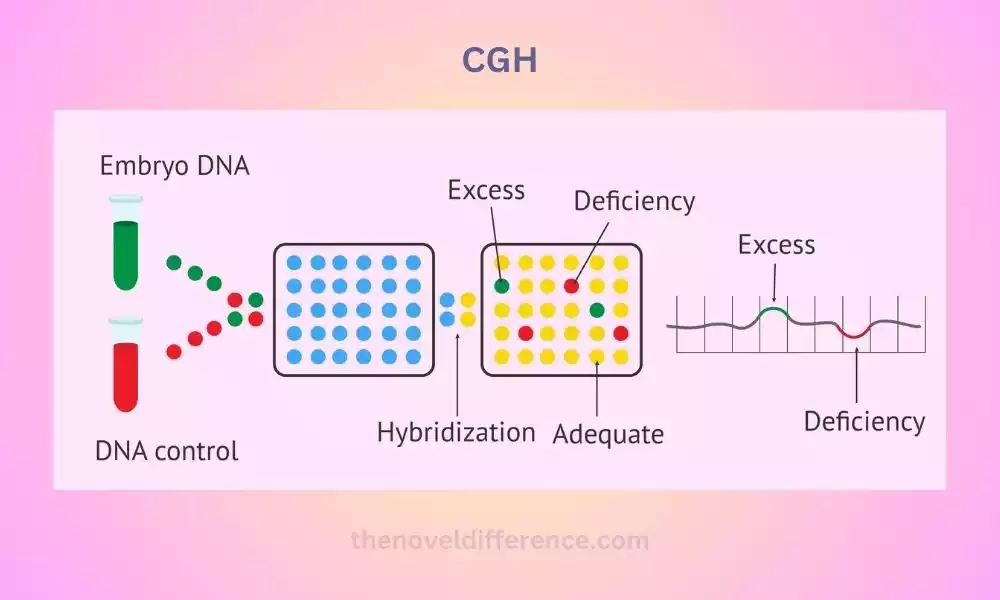
Comparative Genomic Hybridization (CGH) techniques are powerful tools that enable comprehensive analysis of genomic alterations, contributing to our understanding of genetic diseases, cancer development, and personalized medicine.
Array Comparative Genomic Hybridization (Array CGH)
Array Comparative Genomic Hybridization (Array CGH) is an advanced molecular cytogenetic technique that utilizes microarray technology to detect and analyze genomic alterations with high resolution and genome-wide coverage.
It provides a comprehensive assessment of copy number variations (CNVs), gene dosage alterations, and other structural changes in DNA sequences across the entire genome.
A DNA microarray chip is used, consisting of thousands to millions of immobilized genomic DNA fragments or probes. These probes are specifically engineered to target specific regions of the genome – such as known genes, regulatory regions, or functionally significant sequences – using probes designed to target them.
The patient’s DNA and a reference DNA sample (often a healthy individual’s DNA) are labeled with different fluorophores, such as Cy5 and Cy3, respectively.
The labeled patient and reference DNA samples are co-hybridized to the microarray chip, where they competitively bind to the immobilized probes.
Following hybridization, the microarray is scanned to detect the fluorescence intensities of the bound labeled DNA. The fluorescence signals are quantified and analyzed to determine the relative copy number changes between the patient and reference DNA.
By comparing the signal intensities and ratios, Array CGH allows for the identification of gains or losses of genetic material, indicating genomic alterations.
The high density and coverage of probes on the microarray chip enable the detection of small-scale genomic changes with improved resolution compared to traditional CGH methods.
Array CGH offers several advantages over traditional CGH techniques. It provides higher resolution, allowing for the identification of more minor genomic alterations.
EISA also permits simultaneous analysis of multiple loci or genomic regions, providing a thorough assessment of the entire genome in just one experiment. Array CGH has a standardized and automated workflow, enabling higher throughput and improved reproducibility.
Array CGH has been widely used in both research and clinical applications. In research, it is employed to study genomic variations associated with cancer, developmental disorders, genetic syndromes, and other complex diseases.
Array CGH is utilized for diagnostic purposes, enabling the identification of genetic abnormalities and providing valuable information for personalized medicine, genetic counseling, and disease management.
Array CGH has significantly advanced our understanding of genomic alterations and their implications for human health and disease. Genomic sequencing technology has revolutionized genomic analysis by providing more thorough and high-resolution assessments of genomic imbalances.
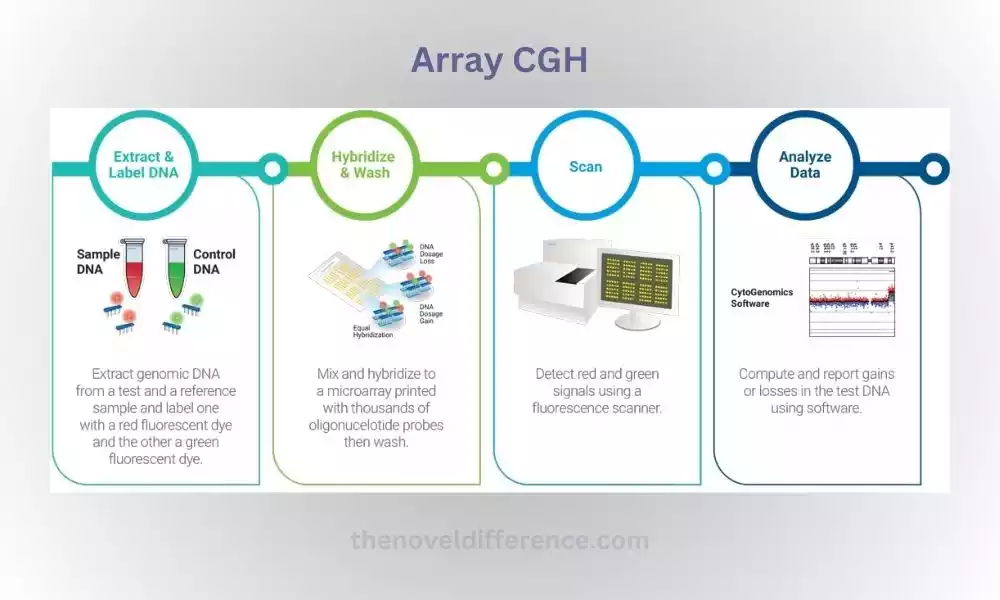
With ongoing advancements in technology, Array CGH continues to contribute to precision medicine, targeted therapies, and the development of innovative diagnostic tools.
Differences Between CGH and Array CGH
CGH and Array CGH (Array Comparative Genomic Hybridization) are two techniques designed to detect and analyze genomic changes; however, each differs significantly in methodology and capabilities.
Here are some key differences between CGH and Array CGH:
- Methodology:
-
- CGH: In CGH, the patient DNA and reference DNA are labeled with different fluorophores and co-hybridized to normal metaphase chromosomes or DNA probes immobilized on slides. The hybridization occurs on a slide with metaphase chromosomes or spotted probes, and the fluorescence signals are visualized and analyzed to detect copy number changes.
- Array CGH: Array CGH utilizes a DNA microarray chip containing thousands to millions of immobilized DNA probes. The patient and reference DNA samples are labeled with different fluorophores and co-hybridized to the microarray. The fluorescence signals emitted by the bound DNA on the microarray are quantified and analyzed to determine copy number changes.
- Resolution and Coverage:
-
- CGH: CGH has a lower resolution compared to Array CGH. It provides a genome-wide assessment of copy number changes but with a limited resolution to detect small-scale alterations.
- Array CGH: Array CGH offers higher resolution due to the high density of probes on the microarray chip. It allows for the detection of small-scale copy number changes and provides more detailed genomic information.
- Throughput and Scalability:
-
- CGH: CGH is generally a low-throughput technique as it involves the analysis of metaphase chromosomes or a limited number of probes.
- Array CGH: Array CGH is a high-throughput technique. It allows for the simultaneous analysis of thousands to millions of probes across the genome, providing a comprehensive assessment of genomic alterations in a single experiment. Array CGH is also scalable, enabling the analysis of larger sample sizes or multiple samples in parallel.
- Cost Considerations:
-
- CGH: CGH generally has lower costs compared to Array CGH. The equipment and reagent requirements are typically less complex and expensive.
- Array CGH: Array CGH can be more costly due to the need for microarray chips and the higher complexity of the technology. The cost of microarray chips and data analysis should be considered.
- Practical Considerations and Sample Requirements:
-
- CGH: CGH requires the availability of metaphase chromosomes, which can be technically challenging to prepare. It may require expertise in cytogenetics.
- Array CGH: Array CGH does not rely on metaphase chromosomes and can be performed using DNA extracted from various sources, including fresh or frozen tissues, blood, or formalin-fixed paraffin-embedded (FFPE) samples. It offers greater flexibility in sample types and is less dependent on sample quality.
It’s worth noting that while Array CGH has several advantages over CGH, the choice between the two techniques depends on the specific research or clinical needs, available resources, and desired resolution. In some cases, CGH may still be preferred for certain applications or when high-resolution analysis is not critical.
Choosing Between CGH and Array CGH
When choosing between CGH and Array CGH techniques, several factors need to be considered based on the specific research or clinical goals, available resources, and desired resolution.
Here are some key considerations to help make a decision:
- Research or Clinical Objectives: Consider the specific goals of the study or analysis. If a broad overview of genomic alterations is sufficient and high resolution is not essential, traditional CGH may be appropriate. However, if a more comprehensive and detailed analysis of copy number changes and higher resolution is required, Array CGH is generally the preferred choice.
- Resolution Requirements: Evaluate the level of resolution needed to detect genomic alterations of interest. Array CGH offers higher resolution due to the dense probe coverage on the microarray chip, enabling the detection of smaller-scale alterations. If the study requires identifying small copy number changes or precise mapping of genomic alterations, Array CGH is recommended.
- Throughput and Sample Size: Assess the number of samples to be analyzed and the desired throughput. Array CGH is a high-throughput technique that allows for the simultaneous analysis of multiple samples in a single experiment. If large sample sizes or parallel analysis of multiple samples is required, Array CGH offers significant advantages in terms of efficiency and scalability.
- Cost Considerations: Consider the available budget and cost constraints. CGH generally has lower costs compared to Array CGH. The equipment and reagent requirements for traditional CGH are typically less complex and expensive. If cost is a major concern, CGH may be a more affordable option.
- Expertise and Resources: Evaluate the available expertise and resources in the laboratory or clinical setting. Traditional CGH requires expertise in cytogenetics and the preparation of metaphase chromosomes, which can be technically challenging. Array CGH, on the other hand, is more accessible and less reliant on cytogenetic expertise. Assess the available infrastructure, equipment, and personnel expertise to determine which technique aligns better with the available resources.
- Sample Types and Compatibility: Consider the types of samples available for analysis. CGH generally requires metaphase chromosomes, while Array CGH can utilize various sample types, including fresh or frozen tissues, blood, or formalin-fixed paraffin-embedded (FFPE) samples. Assess the compatibility of the models with each technique and ensure that the chosen method can accommodate the sample types available.
The choice between CGH and Array CGH depends on the specific requirements of the study, the level of resolution needed, available resources, and budget constraints.
It is advisable to consult with experts in the field, consider the pros and cons of each technique, and weigh them against the specific needs of the research or clinical application to make an informed decision.
Future Directions and Emerging Technologies
The field of genomic analysis, including techniques like CGH and Array CGH, is constantly evolving and advancing. Several future directions and emerging technologies are shaping the landscape of genomic research and clinical applications.
Here are some notable trends and developments to consider:
- Next-Generation Sequencing: NGS technologies such as whole-genome sequence (WGS), and targeted sequencing are becoming more accessible and cost-effective. NGS provides comprehensive genomic information, including copy number variations, structural rearrangements, and sequence variants, in a single experiment. It offers higher resolution and enables simultaneous analysis of multiple genomic alterations. NGS-based approaches are likely to complement or replace traditional CGH and Array CGH methods in the future.
- Single-Cell Genomics: Single-cell genomics techniques allow for the analysis of genomic alterations at the resolution of individual cells. These methods enable the identification and characterization of rare genomic events and intra-tumor heterogeneity. Single-cell CGH and single-cell sequencing technologies provide insights into clonal evolution, cellular diversity, and the impact of genomic alterations on individual cells. They hold great promise for understanding disease progression, treatment response, and personalized medicine.
- Liquid Biopsies: Liquid biopsies involve the analysis of cell-free DNA (cfDNA) or circulating tumor DNA (ctDNA) obtained from blood or other bodily fluids. These noninvasive techniques provide an overview of genomic alterations present in tumors without needing invasive biopsies to analyze them. Liquid biopsies have the potential to revolutionize cancer diagnosis, monitor treatment response, detect minimal residual disease, and identify actionable genomic alterations.
- Artificial Intelligence (AI) and Machine Learning: AI and machine learning algorithms are increasingly being employed to analyze and interpret large-scale genomic data. These computational methods assist in identifying patterns, predicting outcomes, and stratifying patients based on genomic alterations. AI-powered approaches may increase accuracy and efficiency in genomic analysis, providing more precise diagnosis, prognosis, and treatment selection decisions.
- Spatial Genomics: Spatial genomics technologies allow for the visualization and analysis of genomic alterations in their spatial context within tissues. Methods such as spatial transcriptomics, spatial proteomics, and in situ sequencing enable the mapping of genomic alterations to specific cellular and tissue compartments. These techniques provide insights into the spatial heterogeneity of genomic alterations and their influence on tissue organization, cellular interactions, and disease pathology.
- Integration of Multi-Omics Data: The integration of genomic data with other omics data, such as transcriptomics, epigenomics, and proteomics, is gaining importance. Multiple-omics approaches enable an in-depth investigation of genomic changes that alter gene expression, protein activity, and cell processes. Integrative analysis enables the identification of novel biomarkers, therapeutic targets, and molecular signatures associated with the disease.
- Innovation in Data Analysis and Interpretation: Genomic data analysis has evolved rapidly over time due to new algorithms, software tools, and databases being released almost weekly. Efforts are being made to enhance data standardization, integration, and sharing to facilitate collaborative research and clinical applications. User-friendly bioinformatics tools and platforms are being designed to facilitate the analysis and interpretation of genomic data for researchers and clinicians of all levels.
These future directions and emerging technologies hold tremendous potential to advance our understanding of genomic alterations, improve disease diagnosis and treatment, and drive personalized medicine forward.
Continued research and innovation in these areas will shape the future of genomics and contribute to improved patient outcomes.
Conclusion
Comparative Genomic Hybridization (CGH) and Array Comparative Genomic Hybridization (Array CGH) are powerful techniques used to study genomic alterations. CGH provides a global view of copy number changes, while Array CGH offers higher resolution and comprehensive analysis of genomic alterations with its microarray technology.
Studying genomic alterations is of utmost significance as it gives insights into the genetic basis for diseases like cancer and genetic disorders. It helps in understanding disease mechanisms, identifying potential therapeutic targets, guiding treatment decisions, and developing personalized medicine approaches.
