Two common techniques that are frequently employed for various purposes are membrane filtration and direct inoculation. While both methods are used to isolate and identify microorganisms, they differ significantly in their approach and applications.
This article points supply a nitty gritty comparison of the contrasts between layer filtration and coordinate vaccination methods, highlighting their special characteristics, preferences, and impediments.
Definition of Membrane Filtration and Direct Inoculation
Membrane Filtration: Membrane filtration could be a strategy utilized for the partition and concentration of microorganisms from a fluid test. It includes passing the test through a permeable layer channel that holds the microorganisms while permitting the fluid and other smaller particles to pass through. The retained microorganisms on the membrane can then be further processed and analyzed for various purposes, such as microbial enumeration, identification, and characterization.
Direct Inoculation: Direct inoculation, also known as the plate count method or spread plate technique, is a culture-based technique used for the detection and enumeration of microorganisms in a sample. A known volume of the sample is directly inoculated onto a solid culture medium, typically agar, and spread evenly across the surface.
The inoculated plates are then incubated under appropriate conditions to allow the growth of microorganisms. After incubation, visible microbial colonies that develop on the surface of the agar are counted and used to estimate the original microbial population in the sample. This technique relies on the ability of microorganisms to grow and form colonies on the solid medium.
Importance of these techniques in various fields
Membrane filtration and direct inoculation techniques play significant roles in various fields due to their unique capabilities and applications.
Here are some examples of the importance of these techniques in different domains:
1. Food and Beverage Industry:
• Membrane filtration: It is used for the concentration and separation of microorganisms in food and beverage samples, allowing for the detection of potential pathogens and spoilage organisms. It helps in ensuring the safety and quality of food products.
• Direct inoculation: This technique is employed to assess the microbial contamination levels in food and beverage products, aiding in quality control and compliance with regulatory standards.
2. Environmental Monitoring:
• Membrane filtration: It enables the monitoring and analysis of microbial populations in water, wastewater, and environmental samples. It is essential for assessing water quality, identifying pollution sources, and evaluating the effectiveness of water treatment processes.
• Direct inoculation: This method helps in monitoring the microbial contamination of soil, air, and water samples, providing insights into the presence and distribution of microorganisms in various environmental settings.
3. Pharmaceutical and Healthcare Industries:
• Membrane filtration: It is utilized for sterility testing of pharmaceutical products and medical devices. This technique ensures the absence of viable microorganisms, such as bacteria and fungi, in these critical healthcare products.
• Direct inoculation: It is employed for microbial testing of pharmaceutical raw materials, finished products, and clinical samples. This technique assists in evaluating the effectiveness of disinfection and sterilization processes, ensuring product safety and preventing healthcare-associated infections.
4. Research and Laboratory Applications:
• Membrane filtration: It is valuable in research laboratories for studying microbial ecology, investigating microbial diversity, and isolating specific microorganisms of interest from complex samples.
• Direct inoculation: This technique is widely used in microbiology research for studying microbial growth characteristics, antimicrobial susceptibility testing, and screening for novel microorganisms with specific traits or activities.
These techniques are fundamental in many other fields, including biotechnology, water treatment, agriculture, and clinical diagnostics. They provide essential tools for microbial analysis, monitoring, and quality assurance, contributing to the understanding and control of microorganisms in various contexts.
What is Membrane Filtration?
Membrane filtration is a separation technique used to separate and concentrate particles, including microorganisms, from a liquid or gas sample. It involves the use of a porous membrane as a physical barrier that allows the passage of solvent molecules while retaining particles based on their size and molecular properties.
The membrane utilized in filtration is regularly a lean, semi-permeable fabric with few pores. The measure and characteristics of the pores can shift depending on the specified application. Common layer materials incorporate cellulose acetic acid derivation, nitrocellulose, nylon, polyethersulfone, and polyvinylidene fluoride (PVDF).
The process of membrane filtration involves the following steps:
1. Selection of appropriate membrane: The choice of the membrane depends on the nature of the sample, particle size range, and desired separation efficiency.
2. Preparation of the membrane: The membrane is often pre-wetted or soaked in a compatible solution to ensure its integrity and prevent sample loss during filtration.
3. Sample collection and pretreatment: The liquid sample containing the particles or microorganisms of interest is collected and may undergo pretreatment steps such as filtration to remove larger particles or centrifugation to concentrate the sample.
4. Assembly of the filtration setup: The membrane is mounted or placed in a filtration apparatus, such as a filter holder or syringe filter, creating a barrier through which the sample will pass.
5. Filtration process: The sample is applied to the membrane surface and pressure or vacuum is applied to force the liquid through the membrane. The membrane retains particles, including microorganisms, while allowing the liquid phase to pass through.
6. Recovery and analysis of retained microorganisms: After filtration, the retained particles or microorganisms can be recovered from the membrane surface using appropriate techniques such as rinsing with a suitable solvent or using a recovery buffer. These retained microorganisms can then be further analyzed, cultured, enumerated, or characterized as required.
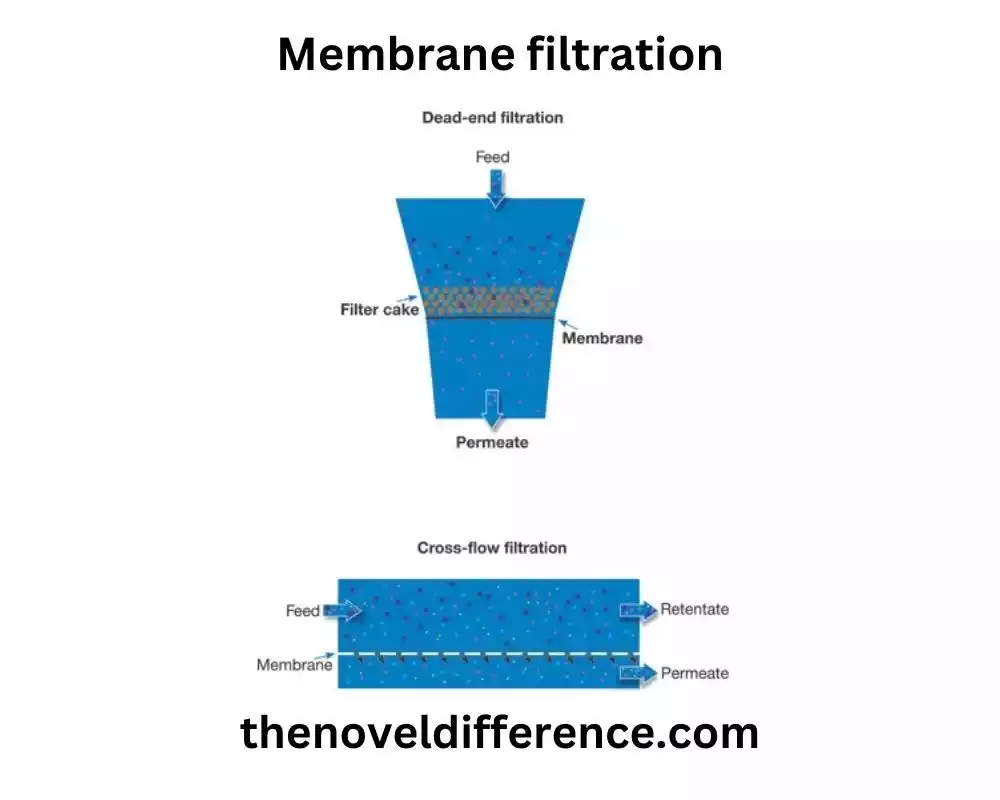
Membrane filtration offers several advantages, including the ability to concentrate microorganisms, flexibility in sample volume processing, efficient removal of particulate matter, and compatibility with various sample types. It finds applications in different areas such as water and wastewater treatment, the nourishment and refreshment industry, pharmaceuticals, natural observing, and investigative research facilities.
Principle of membrane filtration
The principle of membrane filtration is based on the physical separation of particles, including microorganisms, from a liquid or gas sample using a porous membrane. The membrane acts as a selective barrier that allows the passage of the solvent or carrier fluid while retaining particles based on their size and molecular properties.
The key principles underlying membrane filtration are:
1. Size Exclusion: Membranes have microscopic pores of a specific size range. The size of these pores determines the particle size range that can be retained or allowed to pass through. Larger particles, including microorganisms, are hindered by the pores and are retained on the membrane surface, while smaller molecules and solvents can freely pass through.
2. Molecular Sieving: Membranes can also selectively retain particles based on their molecular weight or size. Membranes with specific molecular weight cut-offs (MWCO) are used to separate particles above a certain molecular weight while allowing smaller molecules to pass through.
3. Adsorption and Surface Charge: Membranes can have surface properties that interact with particles. Some membranes possess charged surfaces or specific functional groups that can attract or repel certain particles based on their charge or chemical characteristics. This adsorptive effect can contribute to the retention or passage of particles during filtration.
4. Pressure or Vacuum: The filtration process often involves the application of pressure or vacuum to facilitate the passage of the liquid through the membrane. The applied pressure gradient helps to overcome resistance and drive the liquid flow, enhancing the separation and filtration efficiency.
By utilizing these principles, membrane filtration enables the separation and concentration of microorganisms from a sample, thereby facilitating their subsequent analysis, enumeration, or characterization. The choice of membrane type, pore size, and properties depends on the specific requirements of the application and the size range of particles or microorganisms to be retained or passed through.
Process steps
The process steps involved in membrane filtration are as follows:
1. Selection of appropriate membrane: Choose a membrane with suitable characteristics such as pore size, material composition, and compatibility with the sample and intended analysis. Consider factors like the size range of particles or microorganisms of interest and the nature of the sample matrix.
2. Preparation of the membrane: Pre-wet or soak the membrane in a compatible solution, such as sterile water or buffer, to ensure its integrity and prevent sample loss during filtration. This step helps remove air bubbles and ensures efficient filtration.
3. Sample collection and pretreatment: Collect a representative sample containing the particles or microorganisms you want to separate or concentrate. If necessary, perform pretreatment steps such as filtration to remove larger particles, centrifugation to concentrate the sample, or dilution to adjust the sample volume.
4. Assembly of the filtration setup: Set up the filtration apparatus, which typically includes a filter holder, syringe filter, or filtration unit, according to the manufacturer’s instructions. Ensure a tight and secure connection between the membrane and the filtration device to prevent leakage.
5. Filtration process:
a. Place the prepared membrane in the filtration apparatus, ensuring that the membrane is properly positioned and sealed.
b. Apply pressure or create a vacuum to drive the sample through the membrane. This may be accomplished by employing a vacuum pump, weight filtration framework, or manual weight application, depending on the particular setup.
c. Gradually introduce the sample onto the membrane surface, ensuring a uniform distribution across the membrane.
d. Monitor the filtration process to maintain a steady flow rate, adjusting the pressure or vacuum if necessary. Avoid excessively high pressures that could damage the membrane.
6. Recovery and analysis of retained microorganisms:
a. Once the filtration is complete, carefully remove the membrane from the filtration apparatus.
b. Rinse the membrane with an appropriate solvent or recovery buffer to wash away any loosely retained particles and improve recovery efficiency.
c. Transfer the membrane to a suitable container or culture medium for further analysis or enumeration of the retained microorganisms.
d. Depending on the specific requirements, the retained microorganisms can be further cultured, identified, enumerated, or subjected to additional analysis methods.
It is important to follow good laboratory practices, including proper aseptic techniques and adherence to safety protocols, throughout the membrane filtration process to ensure accurate and reliable results.
Filtration process
The filtration process in membrane filtration involves the following steps:
1. Selection of Appropriate Membrane: Choose a membrane with suitable characteristics such as pore size, material composition, and compatibility with the sample. Consider factors like the size range of particles or microorganisms of interest and the nature of the sample matrix.
2. Preparation of the Membrane: Pre-wet or soak the membrane in a compatible solution, such as sterile water or buffer, to ensure its integrity and prevent sample loss during filtration. This step helps remove air bubbles and ensures efficient filtration.
3. Setup Filtration Apparatus: Assemble the filtration apparatus according to the specific setup being used. This may include a filter holder, syringe filter, or filtration unit. Ensure proper alignment and connection of all components to prevent leakage.
4. Sample Loading: Load the sample onto the membrane surface. This may be done by carefully pouring the sample into the filtration device, employing a syringe to apply the test, or interfacing the device to a collection vessel containing the test.
5. Application of Pressure or Vacuum: Apply pressure or create a vacuum to drive the sample through the membrane. The pressure or vacuum can be generated using a vacuum pump, pressure filtration system, or manual pressure application, depending on the specific setup. Ensure a steady and controlled flow rate to optimize filtration efficiency.
6. Filtration Monitoring: Monitor the filtration process to maintain the desired flow rate and prevent excessive pressure or vacuum that could damage the membrane. Adjust the pressure or vacuum as needed to maintain a consistent flow rate.
7. Completion of Filtration: Continue the filtration preparation until the specified volume of the test has passed through the film or until the required level of molecule or microorganism concentration has been accomplished.
8. Membrane Removal: Once the Filtration is Complete, Carefully Remove the Membrane From the Filtration Apparatus. Handle the membrane with care to avoid contamination or damage.
9. Rinse and Recovery: Rinse the membrane with an appropriate solvent or recovery buffer to wash away any loosely retained particles and improve recovery efficiency. Transfer the membrane to a suitable container or culture medium for further analysis or enumeration of the retained microorganisms.
It is important to take after legitimate aseptic procedures, keep up a clean working environment, and follow security conventions all through the filtration handle to guarantee precise and solid results. Fitting transfer of the utilized filtration device, counting the membrane, ought to be carried out concurring to set up rules and controls.
What is Direct Inoculation?
Direct inoculation, also known as the plate count method or spread plate technique, is a culture-based method used for the detection, enumeration, and isolation of microorganisms in a sample. It involves directly inoculating a known volume of the sample onto a solid culture medium, typically agar, and spreading it evenly across the surface of the medium using a sterile spreader or loop.
The process of direct inoculation includes the following steps:
1. Preparation of Inoculum: Prepare the sample to be tested by collecting a representative portion that contains the microorganisms of interest. The sample can be a liquid, solid, or swabbed from a surface.
2. Inoculation of Sample: Take a known volume of the sample (usually measured in milliliters) and deposit it directly onto the surface of a solid culture medium. This is typically done using a calibrated pipette or other suitable means.
3. Spread Plate Technique: Using a sterile spreader or loop, spread the inoculated sample evenly across the surface of the agar medium. The goal is to distribute the microorganisms uniformly to facilitate their growth and colony formation.
4. Incubation: Place the inoculated agar plate in an incubator set at the appropriate temperature and conditions for the targeted microorganisms to grow. Incubation times can vary depending on the microorganism being targeted.
5. Colony Counting and Identification: After the incubation period, examine the agar plate for visible microbial colonies that have developed. Count the colonies manually or using automated colony counting systems. Each visible colony generally represents a single viable microorganism present in the original sample. Colonies can be further subjected to morphological, biochemical, or molecular tests for identification and characterization.
Direct inoculation is a relatively simple and rapid method for detecting and quantifying viable microorganisms in a sample. It allows for the enumeration of microorganisms and provides valuable information about microbial load and diversity. It is important to note that direct inoculation is limited to culturable microorganisms and does not provide information about viable but non-culturable (VBNC) or unculturable microorganisms.
Principle of direct inoculation
The principle of direct inoculation, also known as the plate count method or spread plate technique, is based on the ability of viable microorganisms to grow and form visible colonies on a solid culture medium.
The principle can be summarized as follows:
1. Dilution and Dispersion: The sample containing microorganisms is diluted to an appropriate concentration, often through serial dilution, to ensure that individual microorganisms are well dispersed in the sample volume.
2. Inoculation: A known volume of the diluted sample is directly inoculated onto the surface of a solid culture medium, typically agar. The inoculum is evenly spread across the medium to ensure uniform distribution of microorganisms.
3. Growth and Colony Formation: The viable microorganisms in the sample, when placed on a suitable nutrient-rich agar medium, have the potential to multiply and form visible colonies. Each colony generally originates from a single viable microorganism present in the inoculated sample.
4. Incubation: The inoculated agar plate is incubated under optimal conditions, including temperature, humidity, and atmospheric composition, for the growth of specific microorganisms of interest. Incubation times vary depending on the microorganism being targeted.
5. Colony Enumeration: After the appropriate incubation period, the visible microbial colonies that have developed on the agar plate are counted. Each counted colony represents a single viable microorganism present in the original sample.
6. Colony Identification and Characterization: Colonies can be further subjected to morphological, biochemical, or molecular tests to identify and characterize the microorganisms. These tests help determine the species or genus of the microorganisms present in the sample.
The principle of direct inoculation relies on the ability of viable microorganisms to grow and form distinct colonies on solid media. By counting and analyzing the colonies, this technique provides valuable information about the viable microbial load, species diversity, and potential contaminants in the sample. It should be noted that direct inoculation is limited to culturable microorganisms and may not detect viable but non-culturable (VBNC) or unculturable microorganisms.
Process steps
The process steps involved in direct inoculation, also known as the plate count method or spread plate technique, are as follows:
1. Sample Collection: Collect a representative sample from the source of interest, such as water, food, soil, or clinical specimens, using appropriate sampling techniques and aseptic precautions.
2. Serial Dilution: Prepare a series of dilutions by transferring a known volume of the sample into a series of sterile dilution tubes containing a suitable diluent, such as sterile saline or buffered peptone water. Typically, a serial dilution is performed to reduce the microbial concentration in the original sample and ensure countable colonies on the agar plate.
3. Inoculation: Take a known volume of a diluted sample (usually measured in milliliters) and transfer it onto the surface of a solid culture medium, such as agar, using a calibrated pipette or a spreader. Place the sample drop on the center of the agar plate.
4. Spread Plate Technique: Using a sterile spreader or loop, spread the inoculated sample evenly over the surface of the agar plate, covering the entire plate or a specific area. Ensure that the sample is uniformly distributed and forms a thin layer on the agar surface.
5. Incubation: Place the inoculated agar plate in an incubator set at the appropriate temperature and conditions required for the growth of the target microorganisms. Incubation times vary depending on the specific microorganisms being targeted, ranging from hours to days.
6. Colony Enumeration: After the incubation period, examine the agar plates for visible microbial colonies that have developed. Count the colonies manually or using automated colony counting systems. Countable colonies are those that are distinguishable from one another and fall within a countable range. It is important to count colonies on plates with suitable colony numbers (typically between 30-300 colonies) to ensure statistical accuracy.
7. Calculation of Colony Forming Units (CFUs): Calculate the colony-forming units per milliliter (CFU/ml) or per gram (CFU/g) of the original sample based on the dilution factor and the number of colonies counted.
8. Identification and Characterization: Depending on the objectives and requirements, selected colonies can be subjected to further identification and characterization using morphological, biochemical, or molecular techniques to determine the species or genus of the microorganisms present.
The direct inoculation method provides a quantitative estimate of viable microorganisms in the sample and allows for the enumeration, identification, and characterization of microbial populations. It could be a broadly utilized procedure in microbiology research facilities for different applications, counting nourishment security, water quality appraisal, clinical microbiology, and nature observing.
Membrane filtration and Direct inoculation
The preparation of the inoculum for direct inoculation involves the following steps:
1. Sample Collection: Collect a representative sample from the source of interest using appropriate sampling techniques and aseptic precautions. The sample can be a liquid, solid, or swabbed from a surface.
2. Sample Homogenization (if applicable): If the sample is non-homogeneous or contains solid particles, it may need to be homogenized to ensure an even distribution of microorganisms. This can be achieved by using a sterile blender, or stomacher, or vortexing the sample with a suitable diluent.
3. Dilution of Sample: Determine the appropriate dilution factor based on the expected microbial concentration in the sample. This step is crucial to obtain countable colonies on the agar plate. Serial weakening is commonly utilized, where aliquots of the test are exchanged into an arrangement of sterile weakening tubes containing a reasonable diluent.
4. Mixing of Diluted Sample: Thoroughly mix each dilution tube by inverting several times or using a vortex mixer to ensure an even distribution of microorganisms throughout the diluent.
5. Inoculation Volume Selection: Decide on the volume of the diluted sample to be inoculated onto the agar plate. The volume will depend on the desired microbial concentration and the countable range of colonies expected on the plate. Usually, volumes between 0.1 mL and 1 mL are used.
6. Inoculation Procedure: Using a calibrated pipette or a loop, transfer the desired volume of the diluted sample onto the surface of the solid culture medium (agar) in a petri dish. Deposit the inoculum in the center of the plate and ensure even spreading by tilting the plate or using a spreader.
7. Incubation: Allow the inoculated agar plates to dry briefly to avoid excessive spreading of the inoculum, and then place them in an incubator set at the appropriate temperature and conditions required for the growth of the target microorganisms. Incubation times may vary depending on the microorganisms of interest.
The proper preparation of the inoculum is crucial to obtain accurate and representative results in the direct inoculation method. It ensures the even distribution of microorganisms on the agar plate, facilitating their growth and colony formation.
Comparison Chart
Here’s a comparison chart highlighting the key differences between membrane filtration and direct inoculation:
| Membrane Filtration | Direct Inoculation |
|---|---|
| Separation of microorganisms based on size | Growth of microorganisms into visible colonies |
| Concentrates microorganisms from a large volume | Does not involve sample concentration |
| More sensitive in detecting low concentrations | Higher detection limit |
| Water analysis, environmental monitoring | Clinical microbiology, food microbiology, monitoring |
| Microorganisms can be recovered from the filter | Visible colonies are counted for further analysis |
| Some microorganisms may not be retained | Limited to viable and culturable microorganisms |
| Does not detect viable but non-culturable | Does not detect viable but non-culturable |
It’s important to note that while this chart provides a general comparison, specific factors may vary depending on the context and specific methodologies used in each technique.
Conclusion
Membrane filtration and Direct inoculation are two distinct techniques used in microbiology laboratories for different purposes. Membrane filtration is suitable for liquid samples and allows the concentration and enumeration of microorganisms, while direct inoculation is used for solid samples to isolate individual colonies. Each strategy has its focal points and drawbacks, and its determination depends on the test sort and the destinations of the investigation. Understanding the differences between membrane filtration and direct inoculation is crucial in choosing the appropriate technique for microbial analysis in various fields.
